Using MLOps to assist with experimentation
In the introduction, we provided some possible outcomes and aspects of the experimentation phase. At the same time, the primary goal of the phase is to find a clear way/solution/algorithm to solve an ML-related problem. Obviously, until we have an understanding of how to solve the problem, we can't start the model development phase, nor build an end-to-end ML flow.
The Importance of MLOps
Some engineers believe that it's a good idea to wait until the experimentation phase is done prior to setting up MLOps. There are two challenges to this thinking:
- The experimentation phase has various outcomes that can be reused in the model development and even in the inferencing phases. Almost all the code, starting from data augmentation up to model training, can be migrated to the end to end ML flow.
- The experimentation phase can be overlapped with the model development phase. The phase begins once we know how to solve the ML problem and now want to tune parameters and components.
These challenges can be summarized into a common problem statement: How can we share experimentation and model development code?
Ignoring this issue at beginning of the project can lead to a situation where data scientists and software engineers are working on two different code bases and spending significant time to sync code between them. The good news is that MLOps provides a way to solve the problem.
MLOps practices to implement
The following sections summarize good MLOps practices that can help data scientists and software engineers collaborate and share code effectively.
Start working with DevOps engineers as soon as possible
Prior to starting any experiment, it's worthwhile to make sure that these conditions have been met:
- Development environment is in place: it can be a compute instance (Azure ML) or an interactive cluster (Databricks) or any other compute (including local one) that allows you to run experiments.
- All data sources should be created: it's a good idea to use an ML platform capability to build technology-specific entities like data assets in Azure ML. It allows you to work with some abstract entities rather than with specific data sources (like Azure Blob).
- Data should be uploaded: it's important to have access to different kinds of data sets to do experiments under different conditions. For example, data might be used on local devices, some may represent a toy data set for validation, and some may represent a full data set for training.
- Authentication should be in place: data scientists should have access to the right data with the ability to avoid entering credentials in their code.
All the conditions are critical and must be managed via collaboration of ML Engineers and DevOps Engineers.
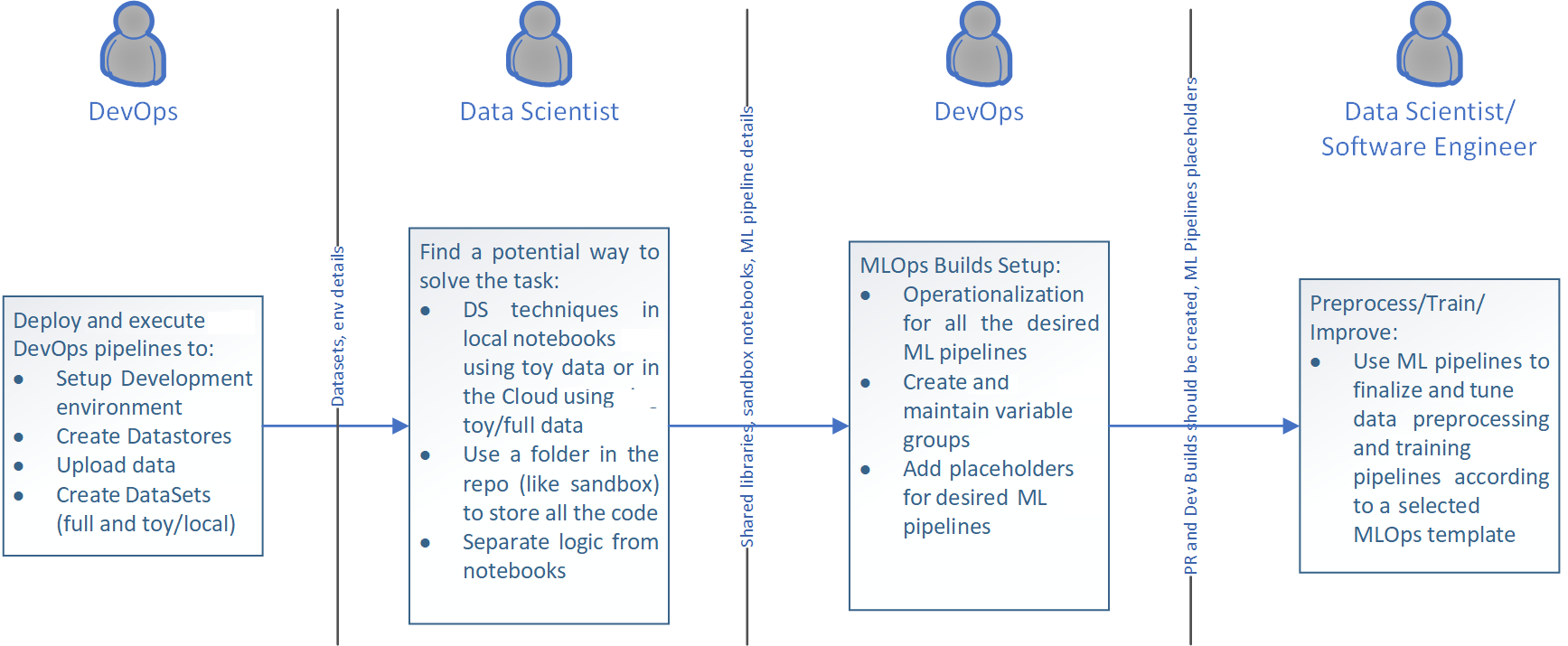
As soon as data scientists have a stable idea, it's important to collaborate with DevOps engineers to make sure all needed code artifacts are in place to start migrating notebooks into pure Python code. The following steps must take place:
- Number of ML flows should be identified: each ML pipeline has its own operationalization aspects and DevOps engineers have to find a way to add all the aspects into CI/CD.
- Placeholders for ML pipelines should be created: each ML pipeline has its own folder structure and ability to use some common code in the repo. A placeholder is a folder with some code and configuration files that represents an empty ML pipeline. Placeholders can be added to CI/CD right away, even prior to migration having been started.
The following diagram shows the stages discussed above:
Start with methods
It doesn't take much effort to wrap your code into methods (even classes) once it's possible. Later, it will be easy to move the methods from the experimentation notebook into pure Python files.
When implementing methods in the experimentation notebook, it's important to make sure that there are no global variables, instead rely on parameters. Later, relying on parameters helps to port code outside of the notebook as is.
Move stable code outside notebooks as soon as possible
If there are some methods/classes to share between notebooks or methods that are not going to be affected much in future iterations, it's worthwhile to move the methods into Python files and import them into the notebook.
At this point, it's important to work with a DevOps engineer to make sure that linting and other checks are in place. If you begin moving code from notebooks to scripts inside the experimentation folder, it is okay to only apply linting. However, if you move your code into a main code folder (like src), it's important to follow all other rules like unit testing code coverage.
Clean notebooks
Cleaning up notebooks before a commit is also good practice. Output information might contain personal information data or non-relevant information that might complicate review. Work with a DevOps engineer to apply some rules and pre-commit hooks. For example, you can use nb-clean.
Executing all notebook cells in sequence should work
Broken notebooks are a common situation when software engineers help data scientists to migrate their notebooks into a pipeline using a selected framework. It's especially true for new ML platforms (Azure ML v2) or for complex frameworks that require much coding to implement a basic pipeline, like Kubeflow. These errors are why it's important that all experimentation notebooks can be executed by a software engineer in order, including all cells, or including clear guidance on why a given cell is optional.
Benefits of using selected ML platform
If you start using Azure ML or a similar technology during the experimentation, the migration will be smooth. For example, the speed benefits gained from using parallel compute in Azure ML may be worth the small amount of additional code required to configure it. Collaboration with Software Engineers can help find some killer feature in the selected ML framework and use it from the beginning.
Next steps
- Experimentation in Azure ML: Different ways to conduct experiments in Azure ML and best practices about how to set up the development environment.