Note
Access to this page requires authorization. You can try signing in or changing directories.
Access to this page requires authorization. You can try changing directories.

Recently at the Microsoft Ignite 2017 conference on the Gold Coast, I gave a talk about some cool new features we’ve introduced in Microsoft R Server 9 in the last 12 months:
- MicrosoftML, a powerful package for machine learning
- Easy deployment of models using SQL Server R Services
- Creating web service APIs with R Server Operationalisation (previously known as DeployR)
To demonstrate these features, as well as showing how Microsoft R interoperates with many open source R packages, I created a sample data science workflow that fits a predictive model, serialises it into SQL Server, and then publishes a web service to get predictions on new data.
In this post, I'll outline the demo workflow, and show some code snippets that give a flavour of what's going on under the hood. The full code is available on GitHub, and you can get it in at either of the following locations:
- https://github.com/Microsoft/acceleratoRs/tree/master/GalaxyClassificationWorkflow
- https://github.com/Microsoft/microsoft-r/tree/master/R%20Server%20Operationalization/GalaxyClassificationWorkflow
You can also see a recording of the Ignite session on Youtube and Channel 9.
Outline of workflow
The problem is derived from the Galaxy Zoo. This is a crowdsourced science project which invites people to classify galaxies using images taken from the Sloan Digital Sky Survey. For the demo, rather than relying on humans, I train a neural network to do the classification instead.
Download the data
The data consists of about 240,000 individual jpeg files (one per galaxy) totalling 2.4GB, along with a catalog table giving basic details on each galaxy. While the original data was obtained from the SDSS website, for the demo I’ve copied it to Azure blob storage.

Some sample images from the Galaxy Zoo
Process the data
The downloaded images are preprocessed before they can be fed into the neural network. Each image is cropped and resized to the same dimensions, and copies are made that are rotated at angles of 45 and 90 degrees. The rotating is because galaxies can appear in photographs at arbitrary angles, and this is an ad-hoc way of making the network insensitive to the angle. The processing is done using the open source imager package, which is a comprehensive suite of tools for image manipulation; and I also use magrittr and dplyr to streamline the data wrangling process.
processImage <- function(angle, files)
{
outPath <- file.path(processedImgPath, angle)
parallel::parSapply(cl, files, function(f, angle, path) {
try(imager::load.image(f) %>%
cropGalaxy(0.26) %>%
rotateGalaxy(angle) %>%
resizeGalaxy(c(50, 50)) %>%
imager::save.image(file.path(path, basename(f))))
},
path=outPath, angle=angle, USE.NAMES=FALSE)
}
# use a parallel backend cluster to speed up preprocessing
cl <- parallel::makeCluster(3)
imgFiles <- dir(imgPath, pattern="\\.jpg$")
lapply(c(0, 45, 90), processImage, files=imgFiles)
Fit the model
Having preprocessed the data, I fit a multilayer convolutional neural network with the rxNeuralNet function from MicrosoftML. This supports an extensive range of network topologies, training algorithms and optimisation hyperparameters. It also supports GPU acceleration via the NVidia CUDA toolkit, which is almost a mandatory requirement when training large networks.
library(MicrosoftML)
# simplified training code
model <- rxNeuralNet(class2 ~ pixels,
data=trainData,
type="multiClass", mlTransformVars="path",
mlTransforms=list(
"BitmapLoaderTransform{col=Image:path}",
"BitmapScalerTransform{col=bitmap:Image width=50 height=50}",
"PixelExtractorTransform{col=pixels:bitmap}"
),
netDefinition=netDefinition,
optimizer=sgd(),
acceleration="gpu",
miniBatchSize=256,
numIterations=100,
normalize="no", initWtsDiameter=0.1, postTransformCache="Disk"
)
rxNeuralNet uses a language called Net# to define the network topology. While you can write Net# code directly (it’s fully documented on MSDN), for convenience, and to avoid having to learn a new language, I’ve used a helper package called RMLtools to generate the code.
Deploy the model
MicrosoftML provides the rxPredict method to compute predicted values, similar to how most other modelling functions in R have a predictmethod. However, this typically only works within R; to do it from outside requires some additional work. Luckily, Microsoft R Server includes some key features to ease this process of model deployment.
Deploying the model in the demo is a two-step process. First, I serialise the model object to a SQL Server table, and create a T-SQL stored procedure that wraps my prediction function.
# simplified prediction code
spBasePredictGalaxyClass <- function(model, imgData)
{
# for each image, preprocess the file and store it
path <- sapply(seq_len(nrow(imgData)), function(i) {
inFile <- tempfile(fileext=".jpg")
outFile <- tempfile(fileext=".jpg")
writeBin(imgData$img[[i]], inFile)
imager::load.image(inFile) %>%
cropGalaxy(0.26) %>%
resizeGalaxy(c(50, 50)) %>%
imager::save.image(outFile)
outFile
})
imgData <- data.frame(galclass=" ", path=path)
model <- rxReadObject(as.raw(model))
rxPredict(model, imgData)
}
library(sqlrutils)
spPredictGalaxyClass <- StoredProcedure(spBasePredictGalaxyClass,
spName="predictGalaxyClass",
InputData("imgData",
defaultQuery="select * from galaxyImgData"),
InputParameter("model", "raw",
defaultQuery="select value from galaxyModels where id='model'"),
connectionString=deployDbConnStr
)
registerStoredProcedure(spPredictGalaxyClass, deployDbConnStr)
Next, I use the mrsdeploy package to publish a web service that can accept new data via an API, and return a predicted class.
apiPredictGalaxyClass <- function(id, img)
{
imgData <- data.frame(specobjid=id, img=img, stringsAsFactors=FALSE)
sqlImgData <- RxSqlServerData("galaxyImgData")
rxDataStep(imgData, sqlImgData, overwrite=TRUE)
sqlrutils::executeStoredProcedure(spPredictGalaxyClass)$data
}
library(mrsdeploy)
apiGalaxyModel <- publishService("apiPredictGalaxyClass", apiPredictGalaxyClass, spPredictGalaxyClass,
inputs=list(id="character", img="character"),
outputs=list(pred="data.frame"))
Consume the model
Finally, I rig up a simple frontend in Shiny that lets you upload new images to send to the neural network to classify. This is an example of using R Server’s built-in tools to consume the API from within R. It's also an example of why you shouldn’t let your backend developers write frontends, but that’s a discussion for another day.
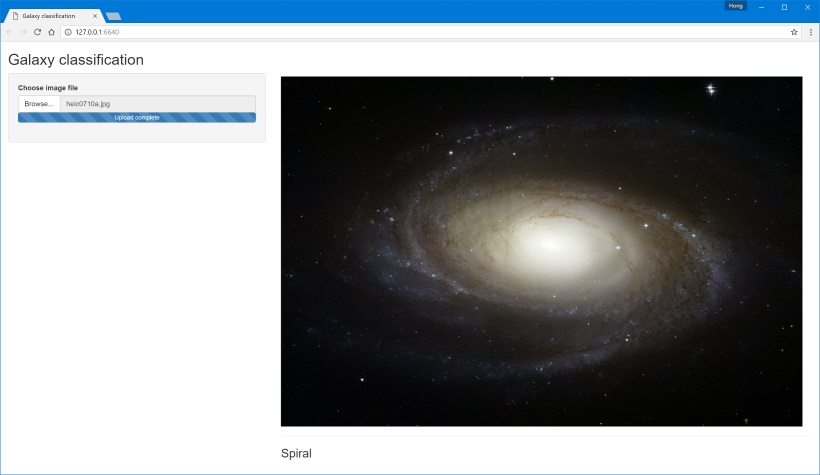
End notes
If you have any comments or questions, please feel free to drop me a line at hongooi@microsoft.com.
Comments
- Anonymous
April 01, 2017
Excellent blog, thanks for sharing.