Understand PCA and import the dataset
Principal component analysis (PCA) is an algorithm that helps us get a dataset into working condition by removing dimensions that must be calculated in an analysis of the dataset.
PCA in theory
One way we reduce the number of dimensions we have to work with is by reducing the number of features considered in an analysis. PCA provides another way: reducing the number of dimensions that we have to work with by projecting our feature space into a lower-dimensional space. We can do this because in most real-world problems, data points aren't spread uniformly across all dimensions. Although some features might be nearly constant, others are highly correlated. The highly correlated data points lie close to a lower-dimensional subspace.
In the following image, the data points aren't spread across the entire plane, but are clumped roughly in an oval. Because the cluster (or any cluster) is roughly elliptical, it can be mathematically described by two values: its major (long) axis and its minor (short) axis. These axes form the principal components of the cluster.
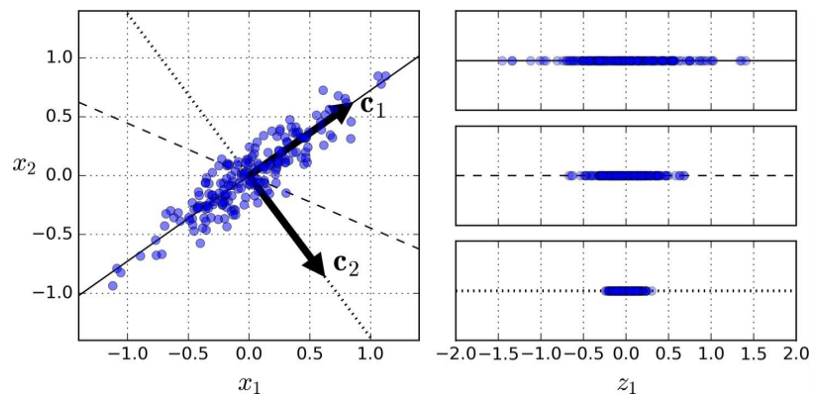
We can construct a whole new feature space around this cluster that's defined by two eigenvectors: $c_{1}$ and $c_{2}$. Eigenvectors are the vectors that define the linear transformation to this new feature space.
Better still, we don't have to consider all of the dimensions of this new space. Intuitively, we can see that most of the points lie on or close to the line that runs through $c_{1}$. If we project the cluster down from two dimensions to that single dimension, we capture most of the information about this dataset while simplifying our analysis. This ability to extract most of the information from a dataset by considering only a fraction of its definitive eigenvectors forms the heart of PCA.
Import modules and dataset
You must first clean and prepare the data to conduct PCA on it, so pandas will be essential. You also need NumPy, a bit of scikit-learn, and pyplot.
To add these libraries, run this code:
import pandas as pd
import numpy as np
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
%matplotlib inline
The dataset we'll use here's the same one that's drawn from the U.S. Department of Agriculture National Nutrient Database for Standard Reference that you prepared in the preceding module.
Remember to set the encoding to latin1 (for µg):
df = pd.read_csv('Data/USDA-nndb-combined.csv', encoding='latin1')
We can check the number of columns and rows by using the info() method for the DataFrame:
df.info()
The output is:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 8989 entries, 0 to 8988
Data columns (total 54 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 NDB_No 8989 non-null int64
1 FoodGroup 8618 non-null object
2 Shrt_Desc 8790 non-null object
3 Water_(g) 8789 non-null float64
4 Energ_Kcal 8790 non-null float64
5 Protein_(g) 8790 non-null float64
6 Lipid_Tot_(g) 8790 non-null float64
7 Ash_(g) 8465 non-null float64
8 Carbohydrt_(g) 8790 non-null float64
9 Fiber_TD_(g) 8196 non-null float64
10 Sugar_Tot_(g) 6958 non-null float64
11 Calcium_(mg) 8442 non-null float64
12 Iron_(mg) 8646 non-null float64
13 Magnesium_(mg) 8051 non-null float64
14 Phosphorus_(mg) 8211 non-null float64
15 Potassium_(mg) 8364 non-null float64
16 Sodium_(mg) 8707 non-null float64
17 Zinc_(mg) 8084 non-null float64
18 Copper_mg) 7533 non-null float64
19 Manganese_(mg) 6630 non-null float64
20 Selenium_(µg) 7090 non-null float64
21 Vit_C_(mg) 7972 non-null float64
22 Thiamin_(mg) 8156 non-null float64
23 Riboflavin_(mg) 8174 non-null float64
24 Niacin_(mg) 8153 non-null float64
25 Panto_Acid_mg) 6548 non-null float64
26 Vit_B6_(mg) 7885 non-null float64
27 Folate_Tot_(µg) 7529 non-null float64
28 Folic_Acid_(µg) 6751 non-null float64
29 Food_Folate_(µg) 7022 non-null float64
30 Folate_DFE_(µg) 6733 non-null float64
31 Choline_Tot_ (mg) 4774 non-null float64
32 Vit_B12_(µg) 7597 non-null float64
33 Vit_A_IU 8079 non-null float64
34 Vit_A_RAE 7255 non-null float64
35 Retinol_(µg) 6984 non-null float64
36 Alpha_Carot_(µg) 5532 non-null float64
37 Beta_Carot_(µg) 5628 non-null float64
38 Beta_Crypt_(µg) 5520 non-null float64
39 Lycopene_(µg) 5498 non-null float64
40 Lut+Zea_ (µg) 5475 non-null float64
41 Vit_E_(mg) 5901 non-null float64
42 Vit_D_µg 5528 non-null float64
43 Vit_D_IU 5579 non-null float64
44 Vit_K_(µg) 5227 non-null float64
45 FA_Sat_(g) 8441 non-null float64
46 FA_Mono_(g) 8124 non-null float64
47 FA_Poly_(g) 8125 non-null float64
48 Cholestrl_(mg) 8380 non-null float64
49 GmWt_1 8490 non-null float64
50 GmWt_Desc1 8491 non-null object
51 GmWt_2 4825 non-null float64
52 GmWt_Desc2 4825 non-null object
53 Refuse_Pct 8740 non-null float64
dtypes: float64(49), int64(1), object(4)
memory usage: 3.7+ MB
Try it yourself
Can you think of a more concise way to check the number of rows and columns in a DataFrame?
Use one of the attributes of the DataFrame.
Here's a possible solution:
df.count(axis='columns')
The output is:
0 54
1 54
2 54
3 54
4 54
..
8984 49
8985 50
8986 53
8987 48
8988 49
Length: 8989, dtype: int64