Deploy and configure DICOM data ingestion in healthcare data solutions (preview)
[This article is prerelease documentation and is subject to change.]
DICOM data ingestion enables you to use the imaging data ingestion pipeline and bring your Digital Imaging and Communications in Medicine (DICOM) data to OneLake. You can deploy and configure the capability after deploying healthcare data solutions (preview) and the Healthcare data foundations capability to your Fabric workspace.
DICOM data ingestion is an optional capability under healthcare data solutions in Microsoft Fabric (preview). You have the flexibility to decide whether or not to use it, depending on your specific needs or scenarios.
Deployment prerequisites
Deploy healthcare data solutions (preview) to your Fabric workspace.
Note
For the DICOM data ingestion imaging capability, you don't need to deploy Azure Health Data Services (FHIR service), the Azure Language service, or the Azure Marketplace offer.
DICOM data ingestion has a direct dependency on the Healthcare data foundations capability. Make sure you successfully deploy, configure, and execute the Healthcare data foundations pipelines first. For more information, see Deploy and configure Healthcare data foundations.
Deploy and configure the OMOP analytics capability (optional).
Download the sample data in your environment as explained in Deploy sample data. If you previously downloaded the sample data, verify whether your download includes the DICOM imaging sample datasets.
Ensure the Fabric runtime version is set to Runtime 1.1 in the workspace settings. For more information, see Reset Spark runtime version in the Fabric workspace.
Configure and execute the following notebooks based on the sample data:
After completing these steps, you should have all the lakehouses deployed and sample clinical datasets ingested in the bronze, silver, and gold delta tables.
To verify the deployment and ingestion of the sample datasets:
- Confirm if you can view the three lakehouses in your workspace window.
- Run a sample query on the care_site table in the OMOP (gold) lakehouse.
Deploy DICOM data ingestion
To deploy the DICOM data ingestion capability to your workspace, follow these steps:
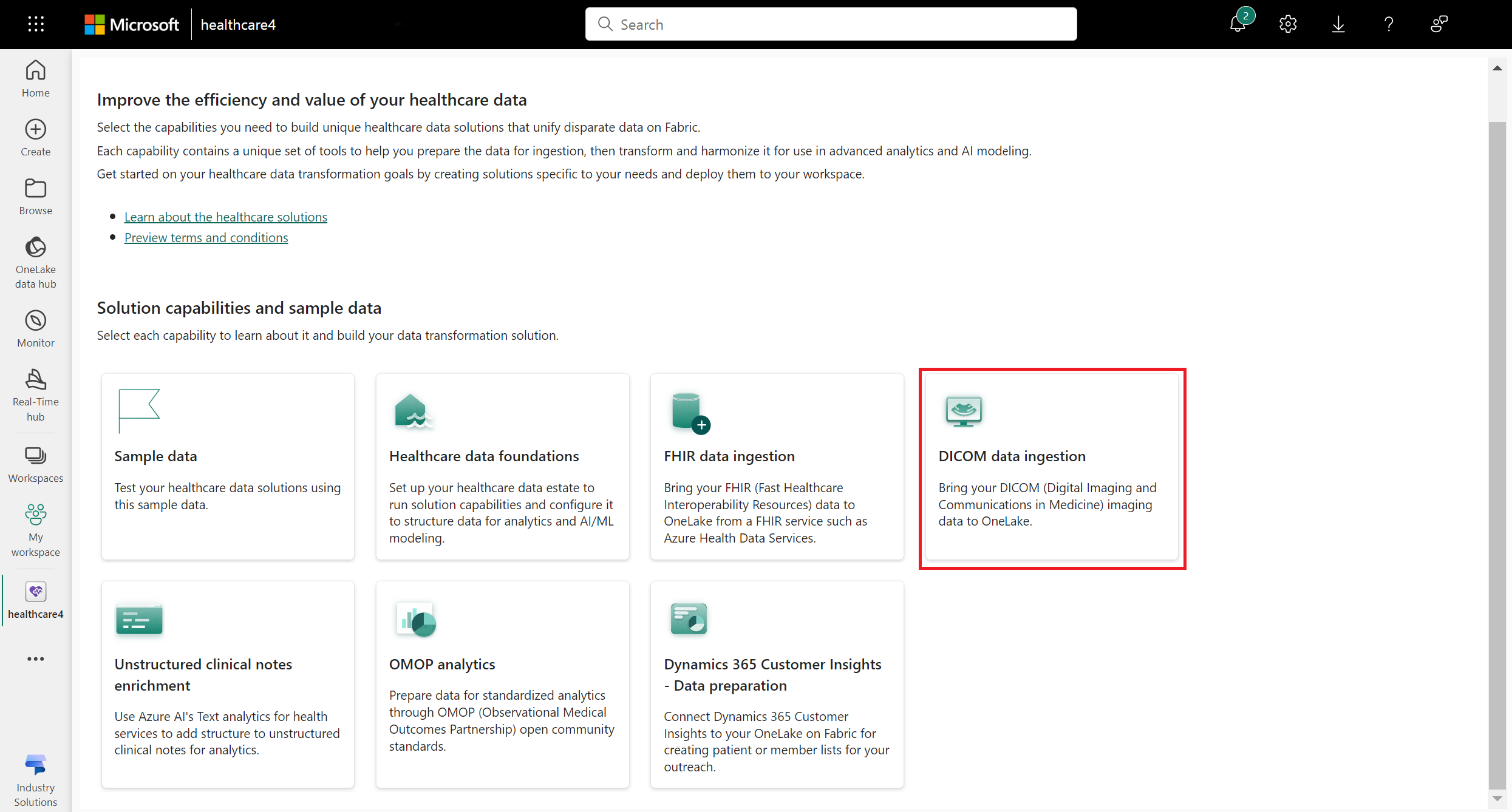
Navigate to the healthcare data solutions home page on Fabric.
Select the DICOM data ingestion tile.
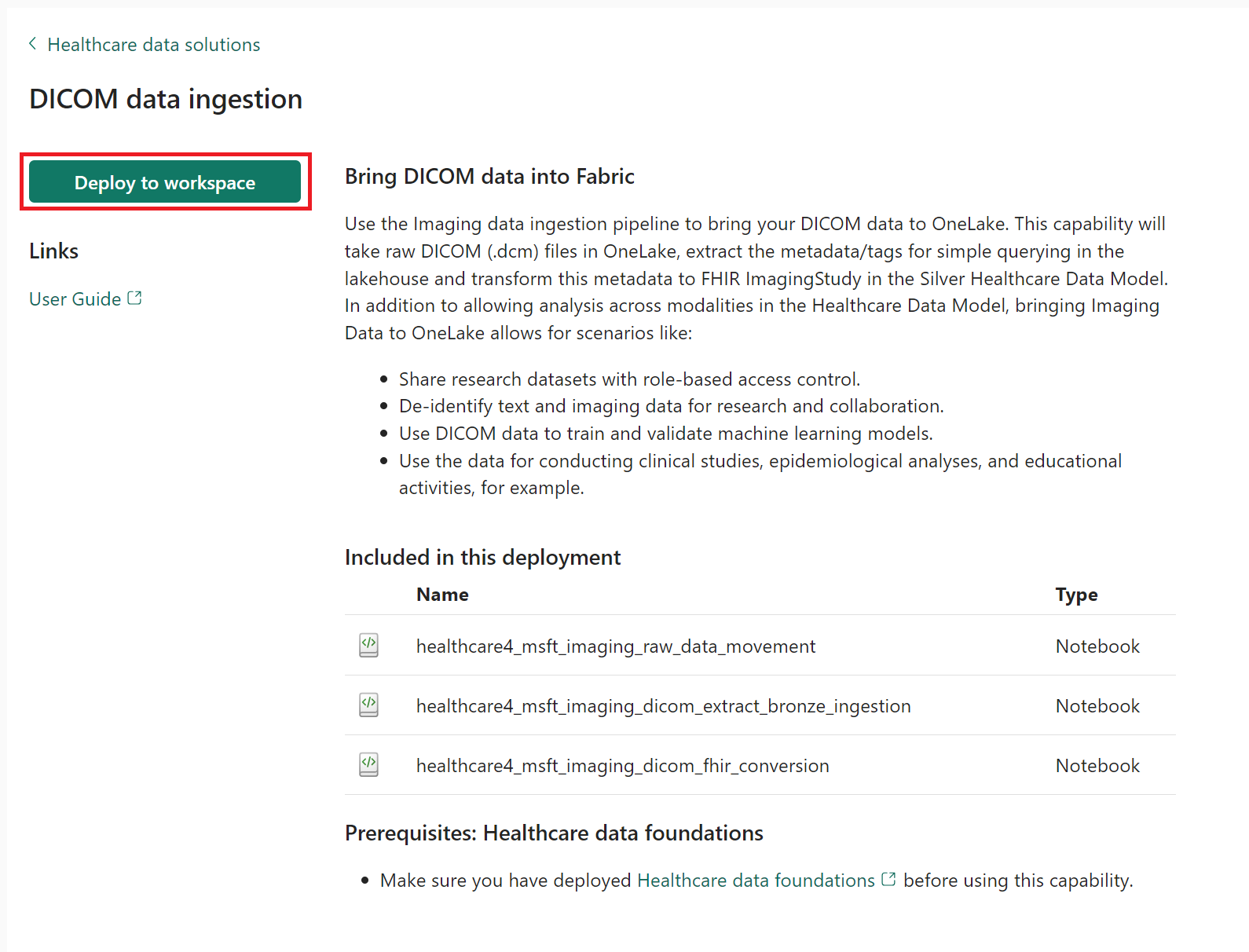
On the capability page, select Deploy to workspace.
The deployment can take a few minutes to complete. Don't close the tab or the browser while deployment is in progress. While you wait, you can work in another tab.
After the deployment completes, you can see a notification on the message bar.
Select Manage capability from the message bar to go to the Capability management page.
Here, you can view, configure, and manage the following three notebooks deployed with the capability:
- healthcare#_msft_imaging_raw_data_movement
- healthcare#_msft_imaging_dicom_extract_bronze_ingestion
- healthcare#_msft_imaging_dicom_fhir_conversion
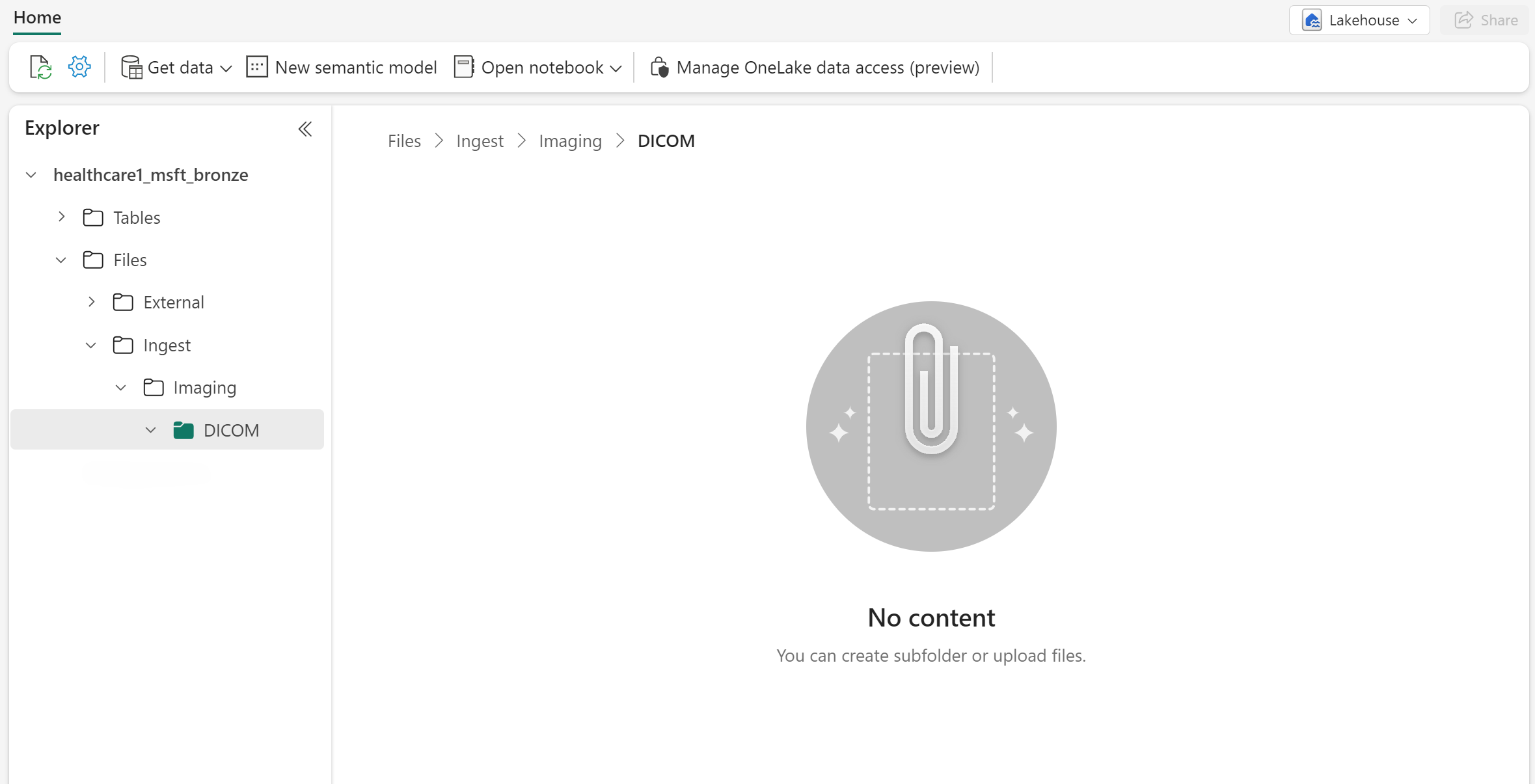
In the same environment, navigate to the healthcare#_msft_bronze lakehouse and select it.
Under Files, create three subfolders using the following folder path:
Files\Ingest\Imaging\DICOM.The Ingest folder should be empty now. It serves as your entry folder to add the DICOM files for ingestion.
DICOM data ingestion notebooks
The DICOM data ingestion capability deploys the following notebooks in your environment. Each notebook includes a set of configurable parameters that you can review and edit on the respective configuration management and setup page.
| Notebook | Functionality |
|---|---|
| healthcare#_msft_imaging_raw_data_movement | Uses the ImagingRawDataMovementService module in the healthcare data solutions (preview) library to move and extract medical imaging files (DCM files) from a ZIP file. The files are transferred from a configurable drop folder to an appropriate folder structure based on the execution date (in yyyy/mm/dd format). |
| healthcare#_msft_imaging_dicom_extract_bronze_ingestion | Uses the MetadataExtractionOrchestrator module in the healthcare data solutions (preview) library to extract the DICOM metadata from DCM files. The metadata is then stored in the dicomimagingmetastore delta table in the bronze lakehouse. |
| healthcare#_msft_imaging_dicom_fhir_conversion | Uses the MetadataToFhirConvertor module in the healthcare data solutions (preview) library to convert the DICOM metadata in the bronze delta table. The conversion process involves transforming metadata from the dicomimagingmetastore table into the FHIR ImagingStudy resource in the FHIR R4.3 format. The output is saved as NDJSON files. |
Imaging sample data
The sample data shipped with healthcare data solutions (preview) includes the imaging sample datasets that you can use to execute the DICOM data ingestion pipeline. You can also explore the data transformation and progression through the medallion bronze, silver, and gold lakehouses. The provided imaging sample data might not be clinically meaningful, but they're technically complete and comprehensive to demonstrate the solution's imaging capabilities. To access the sample data folders, make sure you download the imaging data folder msft_dmh_imaging_data as explained in Deploy sample data.
DICOM folder
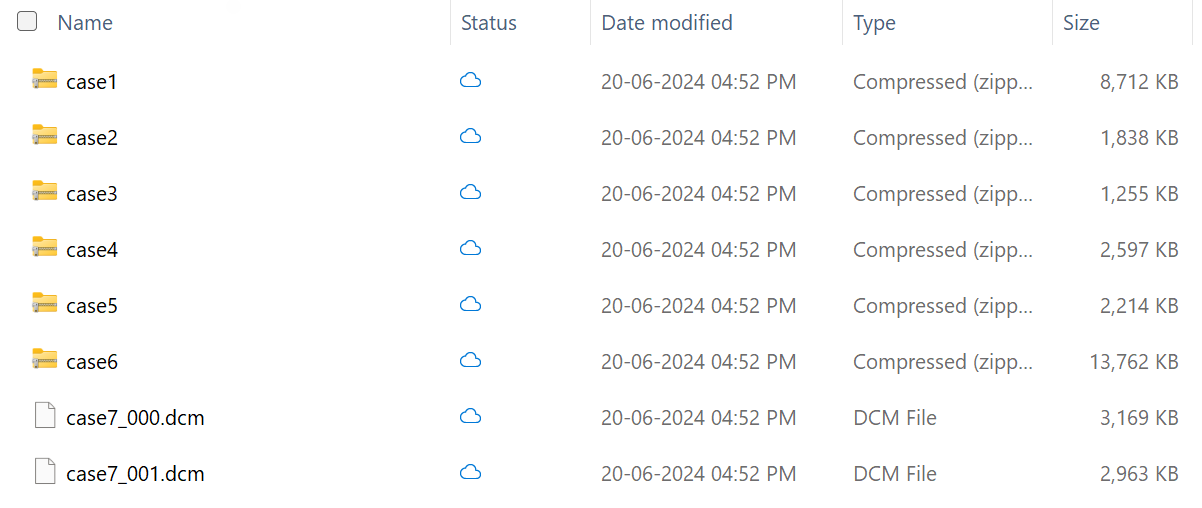
The DICOM folder contains seven case files. The first six files are in ZIP format, while the last file is in the native DCM format. This difference illustrates the ingestion pipeline's ability to ingest DICOM data in both compressed and native formats. The patient identifiers in these imaging case files correspond to the patients in the clinical sample data.
You can use OneLake file explorer or Azure Storage Explorer to access the deployed sample data.
Here's a detailed overview showing the total number of studies (9), series (24), and instances (96) in the imaging dataset:
| Case file | Study description (StudyInstanceUID) | Series (modalities) | Instances |
|---|---|---|---|
| case1.zip | MRT Sakroiliakalgelenke (1.2.276.0.50.192168001099.7810872.14547392.270) | 1 (MR) | 31 |
| case2.zip | CT Abdomen (1.2.276.0.50.192168001099.8252157.14547392.4) | 2 (CT) | 7 |
| case3.zip | MRT Oberbauch (1.2.276.0.50.192168001092.11156604.14547392.4) | 5 (MR) | 7 |
| case4.zip | MRT Schädel (1.2.276.0.50.192168001099.8687553.14547392.4) | 4 (MR) | 30 |
| case5.zip | MRA (1.2.276.0.50.192168001092.11517584.14547392.4) MRT Oberbauch (1.2.276.0.50.192168001099.8829267.14547392.4) | 1 (OT) 5 (MR) | 5 6 |
| case6.zip | Thorax digital (1.2.276.0.50.192168001099.9140875.14547392.277) CT Thorax (1.2.276.0.50.192168001099.9140875.14547392.4) | 2 (CR) 2 (CT) | 2 6 |
| case7.dcm | Ellenbogen (1.2.276.0.50.192168001099.9483698.14547392.4) | 2 (CR) | 2 |
| Total | 9 | 24 | 96 |
where:
- CR = Computed Radiography
- CT = Computerized Tomography
- MR/MRT = Magnetic Resonance Tomography, commonly known as Magnetic Resonance Imaging (MRI)
- MRA = Magnetic Resonance Angiography
- OT = Optical Tomography
Note
Some study descriptions in the previous table are in the German language since the metadata of the sample imaging dataset are in German. Here are the corresponding English translations for these terms:
| German | English |
|---|---|
| Ellenbogen | Elbow |
| Oberbauch | Upper abdomen |
| Sakroiliakalgelenke | Sacroiliac joints |
| Schädel | Skull |
FHIR folder
Important
There's a known issue with the diagnostic reports in the sample data. The diagnostic reports are currently deployed in JSON format instead of NDJSON format. You can't ingest these files as-is; they must be converted to NDJSON first. We'll work on resolving this issue in the next release.
The FHIR folder includes six NDJSON files containing sample DiagnosticReport FHIR resources for the first six DICOM case files and their corresponding patients. You don't need to ingest these diagnostic reports for running the DICOM ingestion pipeline, as the reports are considered clinical data and are ingested as part of FHIR data ingestion. This use case demonstrates multi-model capabilities in healthcare data solutions (preview) that showcase the ability to ingest both clinical and imaging data, offering a realistic and cohesive scenario.
Deploy the DICOM API in Azure Health Data Services
Important
Follow this deployment section only if you're using the DICOM service in Azure Health Data Services. Otherwise, you can skip this section and proceed to Configure DICOM data ingestion.
Azure Health Data Services is a cloud-based solution that helps you collect, store, and analyze health data from different sources and formats. It supports various healthcare standards, such as DICOM. The DICOM service (part of Azure Health Data Services) is a cloud-based solution that enables healthcare organizations to store, manage, and exchange medical imaging data securely and efficiently with any DICOM web-enabled systems or applications.
DICOM data ingestion in healthcare data solutions (preview) has a native integration with the DICOM service. If you're already using the DICOM service, you can also use the imaging analytical capabilities in healthcare data solutions (preview). This native integration eliminates the need for manual integration of datasets between the two services. The integration is based on providing a OneLake shortcut to Azure Data Lake Storage Gen2 for the Azure Health Data Services DICOM service. To set up the data lake integration, follow the steps in Deploy the DICOM service with Azure Data Lake Storage. For more information on this ingestion pipeline, go to Option 3: End to end integration with the DICOM service.
Configure DICOM data ingestion
Configuring the DICOM data ingestion capability involves two levels of configuration in your healthcare data solutions (preview) environment: global configuration and notebook-level configuration.
Global configuration
You can configure the healthcare#_msft_config_notebook notebook to set up and manage the configuration needed for all the data transformations in your healthcare data solutions (preview) environment. For more information, go to Configure the global configuration notebook.
Notebook-level configuration
The ingestion capability deploys the three notebooks listed in DICOM data ingestion notebooks. Some of the notebook configuration parameters are inherited from the global configuration and can be overridden at the notebook level. The notebooks also include specific configuration parameters defined under the kwargs dictionary. You can provide these parameters when you invoke the service library in the code within the notebook.
Configure data ingestion from the FHIR ImagingStudy files
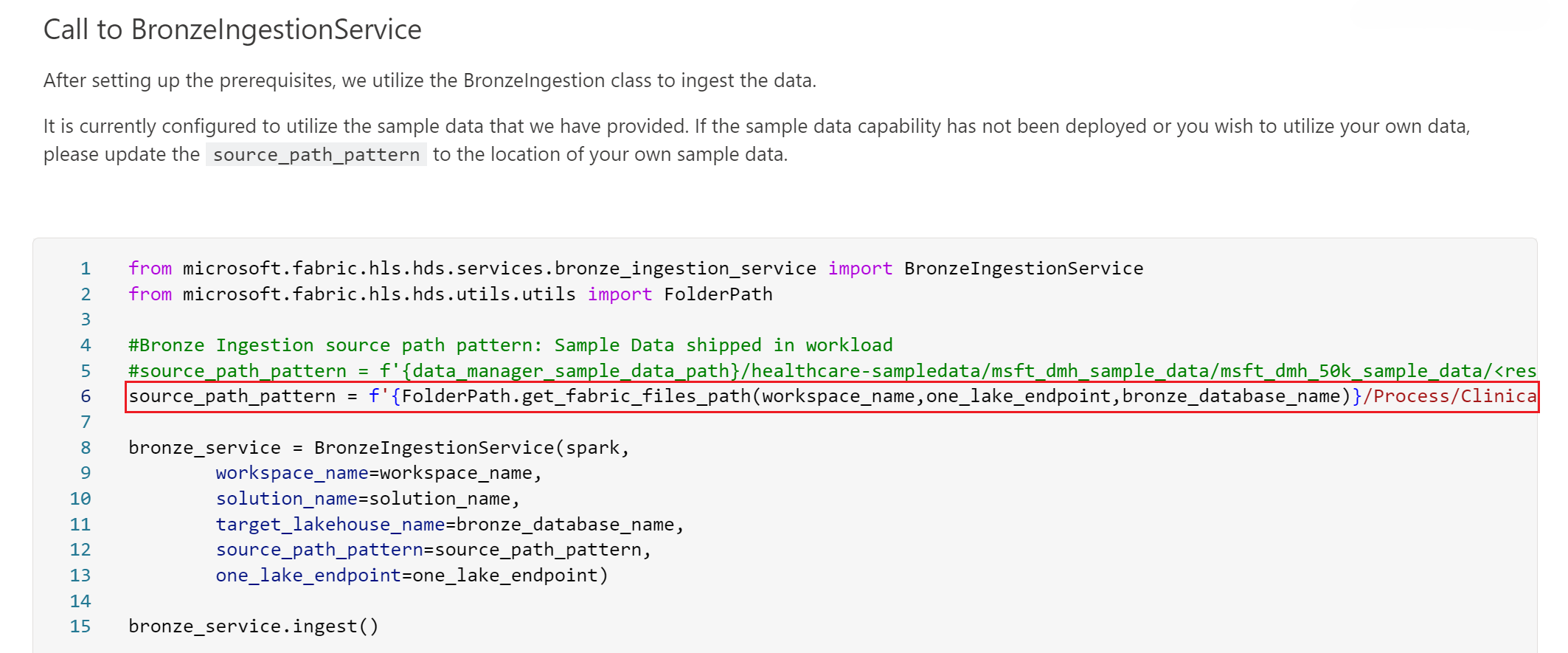
To configure data ingestion from the FHIR ImagingStudy files, you need to modify the source_path_pattern parameter value in the healthcare#_msft_raw_bronze_ingestion notebook. This modification is needed since the preconfigured value for this parameter refers to the clinical sample data folder.
Reconfigure this parameter value to refer to the optimized folder structure for your medical imaging data under Files\Process\Clinical\FHIR NDJSON\Fabric.HDS\yyyy\mm\dd\ImagingStudy. For example, your source_path_pattern parameter might look similar to the following value:
source_path_pattern = f'{FolderPath.get_fabric_files_path(workspace_name,one_lake_endpoint,bronze_database_name)}/Process/Clinical/FHIR NDJSON/Fabric.HDS/**/**/**/ImagingStudy/<resource_name>*ndjson'
Configure Azure Data Lake Storage ingestion
Follow this Bring Your Own Storage (BYOS) configuration option if you wish to use DICOM files from your Azure Data Lake Storage Gen2 storage location. With this option, you don't need to copy or move the DICOM files to the OneLake ingestion folders. Instead, you can create a OneLake shortcut to Data Lake Storage Gen2 and access the DICOM files from their original storage location. To review the detailed steps and the execution pipeline, go to Option 2: DICOM data ingestion from Azure Data Lake Storage Gen2.
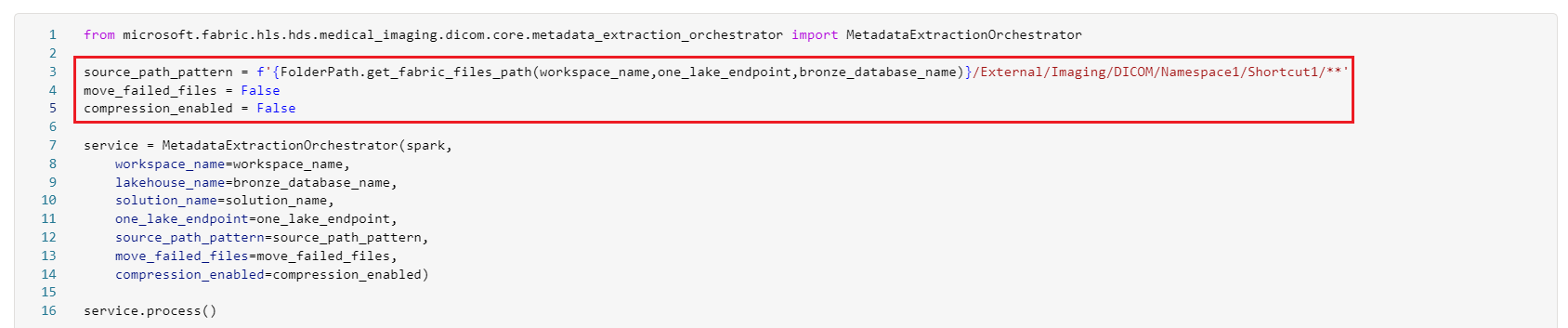
To set up this ingestion option, reconfigure the healthcare#_msft_imaging_dicom_extract_bronze_ingestion notebook parameters. This reconfiguration disables compression or file movement to the failed folders and repoints the notebook to the shortcut path under Files\External\Imaging\DICOM\[Namespace]\[ShortcutName].
Reconfigure the notebook parameters as follows:
source_path_pattern = f'{FolderPath.get_fabric_files_path(workspace_name,one_lake_endpoint,bronze_database_name)}/External/Imaging/DICOM/Namespace1/Shortcut1/**move_failed_files = Falsecompression_enabled = False
Configure data ingestion from the FHIR DiagnosticReport files
Important
There's a known issue with the diagnostic reports in the sample data. The diagnostic reports are currently deployed in JSON format instead of NDJSON format. You can't ingest these files as-is; they must be converted to NDJSON first. We'll work on resolving this issue in the next release.
To ingest the FHIR DiagnosticReport NDJSON files into the bronze lakehouse, modify the source_path_pattern parameter in the healthcare#_msft_raw_bronze_ingestion notebook. Update the parameter value to point to the optimized folder structure for the diagnostic reports under Files\Process\Clinical\FHIR NDJSON\Fabric.HDS\yyyy\mm\dd\DiagnosticReport.
For example, your source_path_pattern parameter might look similar to the following value:
source_path_pattern = f'{FolderPath.get_fabric_files_path(workspace_name,one_lake_endpoint,bronze_database_name)}/Process/Clinical/FHIR NDJSON/Fabric.HDS/**/**/**/DiagnosticReport/<resource_name>*ndjson