Note
Access to this page requires authorization. You can try signing in or changing directories.
Access to this page requires authorization. You can try changing directories.
This article describes how to use the Execute R Script component to run R code in your Azure Machine Learning designer pipeline.
With R, you can do tasks that aren't supported by existing components, such as:
- Create custom data transformations
- Use your own metrics to evaluate predictions
- Build models using algorithms that aren't implemented as standalone components in the designer
R version support
Azure Machine Learning designer uses the CRAN (Comprehensive R Archive Network) distribution of R. The currently used version is CRAN 3.5.1.
Supported R packages
The R environment is preinstalled with more than 100 packages. For a complete list, see the section Preinstalled R packages.
You can also add the following code to any Execute R Script component, to see the installed packages.
azureml_main <- function(dataframe1, dataframe2){
print("R script run.")
dataframe1 <- data.frame(installed.packages())
return(list(dataset1=dataframe1, dataset2=dataframe2))
}
Note
If your pipeline contains multiple Execute R Script components that need packages that aren't in the preinstalled list, install the packages in each component.
Installing R packages
To install additional R packages, use the install.packages() method. Packages are installed for each Execute R Script component. They aren't shared across other Execute R Script components.
Note
It's NOT recommended to install R package from the script bundle. It's recommended to install packages directly in the script editor.
Specify the CRAN repository when you're installing packages, such as install.packages("zoo",repos = "https://cloud.r-project.org").
Warning
Excute R Script component does not support installing packages that require native compilation, like qdap package which requires JAVA and drc package which requires C++. This is because this component is executed in a pre-installed environment with non-admin permission.
Do not install packages which are pre-built on/for Windows, since the designer components are running on Ubuntu. To check whether a package is pre-built on windows, you could go to CRAN and search your package, download one binary file according to your OS, and check Built: part in the DESCRIPTION file. Following is an example:
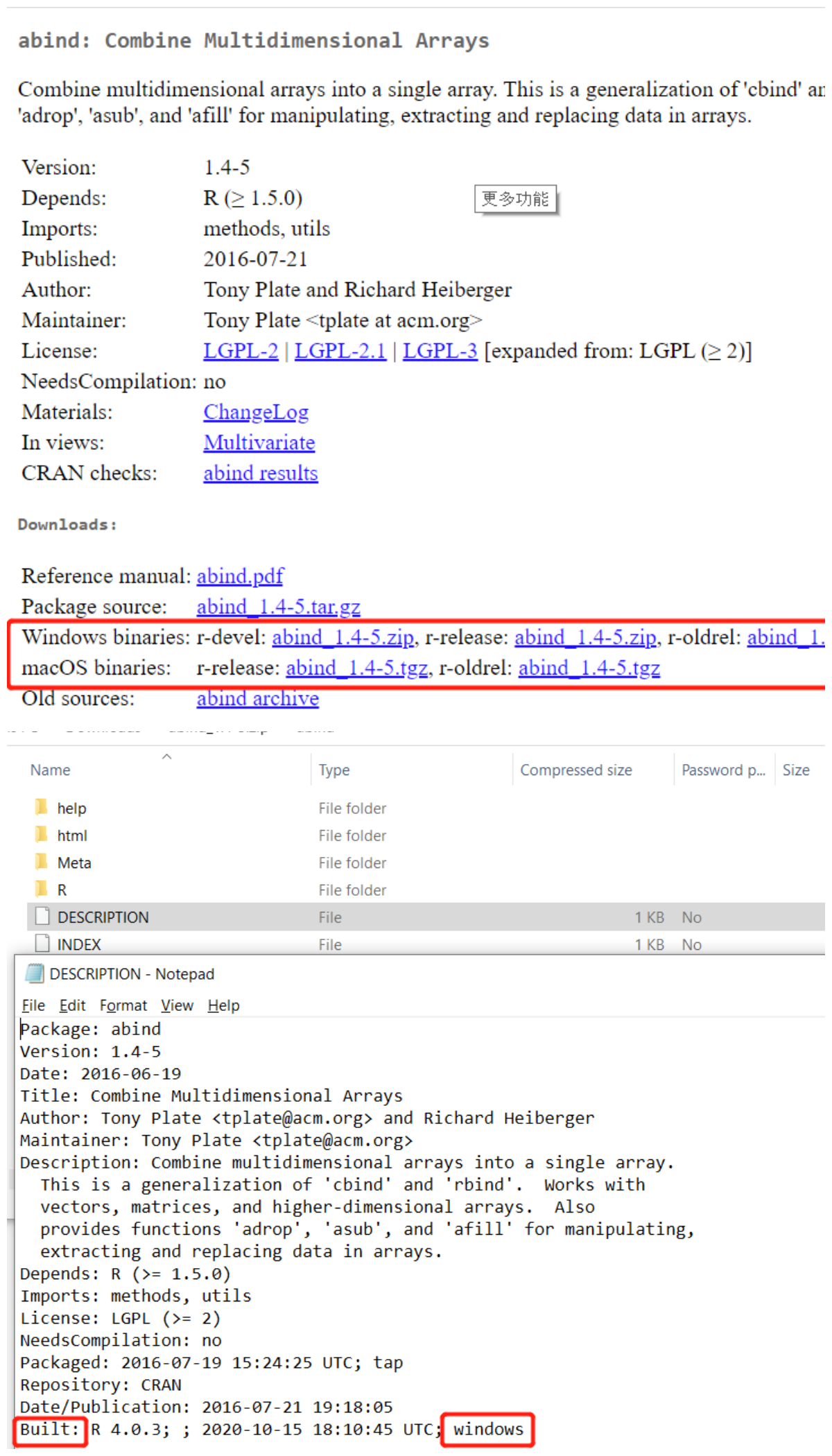
This sample shows how to install Zoo:
# R version: 3.5.1
# The script MUST contain a function named azureml_main,
# which is the entry point for this component.
# Note that functions dependent on the X11 library,
# such as "View," are not supported because the X11 library
# is not preinstalled.
# The entry point function MUST have two input arguments.
# If the input port is not connected, the corresponding
# dataframe argument will be null.
# Param<dataframe1>: a R DataFrame
# Param<dataframe2>: a R DataFrame
azureml_main <- function(dataframe1, dataframe2){
print("R script run.")
if(!require(zoo)) install.packages("zoo",repos = "https://cloud.r-project.org")
library(zoo)
# Return datasets as a Named List
return(list(dataset1=dataframe1, dataset2=dataframe2))
}
Note
Before you install a package, check if it already exists so you don't repeat an installation. Repeat installations might cause web service requests to time out.
Access to registered dataset
You can refer to the following sample code to access to the registered datasets in your workspace:
azureml_main <- function(dataframe1, dataframe2){
print("R script run.")
run = get_current_run()
ws = run$experiment$workspace
dataset = azureml$core$dataset$Dataset$get_by_name(ws, "YOUR DATASET NAME")
dataframe2 <- dataset$to_pandas_dataframe()
# Return datasets as a Named List
return(list(dataset1=dataframe1, dataset2=dataframe2))
}
How to configure Execute R Script
The Execute R Script component contains sample code as a starting point.
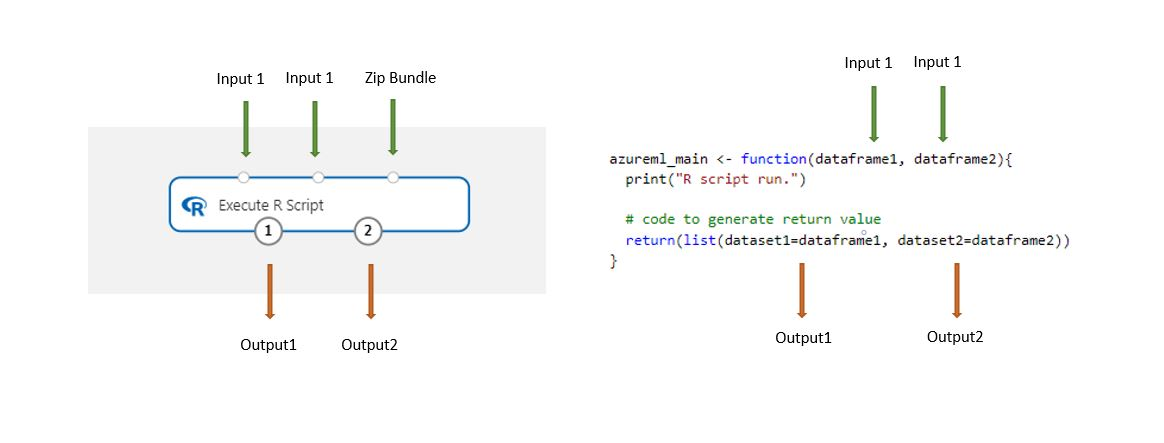
Datasets stored in the designer are automatically converted to an R data frame when loaded with this component.
Add the Execute R Script component to your pipeline.
Connect any inputs that the script needs. Inputs are optional and can include data and additional R code.
Dataset1: Reference the first input as
dataframe1. The input dataset must be formatted as a CSV, TSV, or ARFF file. Or you can connect an Azure Machine Learning dataset.Dataset2: Reference the second input as
dataframe2. This dataset also must be formatted as a CSV, TSV, or ARFF file, or as an Azure Machine Learning dataset.Script Bundle: The third input accepts .zip files. A zipped file can contain multiple files and multiple file types.
In the R script text box, type or paste valid R script.
Note
Be careful when writing your script. Make sure there are no syntax errors, such as using undeclared variables or unimported components or functions. Pay extra attention to the preinstalled package list at the end of this article. To use packages that aren't listed, install them in your script. An example is
install.packages("zoo",repos = "https://cloud.r-project.org").To help you get started, the R Script text box is prepopulated with sample code, which you can edit or replace.
# R version: 3.5.1 # The script MUST contain a function named azureml_main, # which is the entry point for this component. # Note that functions dependent on the X11 library, # such as "View," are not supported because the X11 library # is not preinstalled. # The entry point function MUST have two input arguments. # If the input port is not connected, the corresponding # dataframe argument will be null. # Param<dataframe1>: a R DataFrame # Param<dataframe2>: a R DataFrame azureml_main <- function(dataframe1, dataframe2){ print("R script run.") # If a .zip file is connected to the third input port, it's # unzipped under "./Script Bundle". This directory is added # to sys.path. # Return datasets as a Named List return(list(dataset1=dataframe1, dataset2=dataframe2)) }The entry point function must have the input arguments
Param<dataframe1>andParam<dataframe2>, even when these arguments aren't used in the function.Note
The data passed to the Execute R Script component is referenced as
dataframe1anddataframe2, which is different from Azure Machine Learning designer (the designer reference asdataset1,dataset2). Make sure that input data is referenced correctly in your script.Note
Existing R code might need minor changes to run in a designer pipeline. For example, input data that you provide in CSV format should be explicitly converted to a dataset before you can use it in your code. Data and column types used in the R language also differ in some ways from the data and column types used in the designer.
If your script is larger than 16 KB, use the Script Bundle port to avoid errors like CommandLine exceeds the limit of 16597 characters.
- Bundle the script and other custom resources to a zip file.
- Upload the zip file as a File Dataset to the studio.
- Drag the dataset component from the Datasets list in the left component pane in the designer authoring page.
- Connect the dataset component to the Script Bundle port of Execute R Script component.
Following is the sample code to consume the script in the script bundle:
azureml_main <- function(dataframe1, dataframe2){ # Source the custom R script: my_script.R source("./Script Bundle/my_script.R") # Use the function that defined in my_script.R dataframe1 <- my_func(dataframe1) sample <- readLines("./Script Bundle/my_sample.txt") return (list(dataset1=dataframe1, dataset2=data.frame("Sample"=sample))) }For Random Seed, enter a value to use inside the R environment as the random seed value. This parameter is equivalent to calling
set.seed(value)in R code.Submit the pipeline.
Results
Execute R Script components can return multiple outputs, but they must be provided as R data frames. The designer automatically converts data frames to datasets for compatibility with other components.
Standard messages and errors from R are returned to the component's log.
If you need to print results in the R script, you can find the printed results in 70_driver_log under the Outputs+logs tab in the right panel of the component.
Sample scripts
There are many ways to extend your pipeline by using custom R scripts. This section provides sample code for common tasks.
Add an R script as an input
The Execute R Script component supports arbitrary R script files as inputs. To use them, you must upload them to your workspace as part of the .zip file.
To upload a .zip file that contains R code to your workspace, go to the Datasets asset page. Select Create dataset, and then select From local file and the File dataset type option.
Verify that the zipped file appears in My Datasets under the Datasets category in the left component tree.
Connect the dataset to the Script Bundle input port.
All files in the .zip file are available during pipeline run time.
If the script bundle file contained a directory structure, the structure is preserved. But you must alter your code to prepend the directory ./Script Bundle to the path.
Process data
The following sample shows how to scale and normalize input data:
# R version: 3.5.1
# The script MUST contain a function named azureml_main,
# which is the entry point for this component.
# Note that functions dependent on the X11 library,
# such as "View," are not supported because the X11 library
# is not preinstalled.
# The entry point function MUST have two input arguments.
# If the input port is not connected, the corresponding
# dataframe argument will be null.
# Param<dataframe1>: a R DataFrame
# Param<dataframe2>: a R DataFrame
azureml_main <- function(dataframe1, dataframe2){
print("R script run.")
# If a .zip file is connected to the third input port, it's
# unzipped under "./Script Bundle". This directory is added
# to sys.path.
series <- dataframe1$width
# Find the maximum and minimum values of the width column in dataframe1
max_v <- max(series)
min_v <- min(series)
# Calculate the scale and bias
scale <- max_v - min_v
bias <- min_v / dis
# Apply min-max normalizing
dataframe1$width <- dataframe1$width / scale - bias
dataframe2$width <- dataframe2$width / scale - bias
# Return datasets as a Named List
return(list(dataset1=dataframe1, dataset2=dataframe2))
}
Read a .zip file as input
This sample shows how to use a dataset in a .zip file as an input to the Execute R Script component.
- Create the data file in CSV format, and name it mydatafile.csv.
- Create a .zip file and add the CSV file to the archive.
- Upload the zipped file to your Azure Machine Learning workspace.
- Connect the resulting dataset to the ScriptBundle input of your Execute R Script component.
- Use the following code to read the CSV data from the zipped file.
azureml_main <- function(dataframe1, dataframe2){
print("R script run.")
mydataset<-read.csv("./Script Bundle/mydatafile.csv",encoding="UTF-8");
# Return datasets as a Named List
return(list(dataset1=mydataset, dataset2=dataframe2))
}
Replicate rows
This sample shows how to replicate positive records in a dataset to balance the sample:
azureml_main <- function(dataframe1, dataframe2){
data.set <- dataframe1[dataframe1[,1]==-1,]
# positions of the positive samples
pos <- dataframe1[dataframe1[,1]==1,]
# replicate the positive samples to balance the sample
for (i in 1:20) data.set <- rbind(data.set,pos)
row.names(data.set) <- NULL
# Return datasets as a Named List
return(list(dataset1=data.set, dataset2=dataframe2))
}
Pass R objects between Execute R Script components
You can pass R objects between instances of the Execute R Script component by using the internal serialization mechanism. This example assumes that you want to move the R object named A between two Execute R Script components.
Add the first Execute R Script component to your pipeline. Then enter the following code in the R Script text box to create a serialized object
Aas a column in the component's output data table:azureml_main <- function(dataframe1, dataframe2){ print("R script run.") # some codes generated A serialized <- as.integer(serialize(A,NULL)) data.set <- data.frame(serialized,stringsAsFactors=FALSE) return(list(dataset1=data.set, dataset2=dataframe2)) }The explicit conversion to integer type is done because the serialization function outputs data in the R
Rawformat, which the designer doesn't support.Add a second instance of the Execute R Script component, and connect it to the output port of the previous component.
Type the following code in the R Script text box to extract object
Afrom the input data table.azureml_main <- function(dataframe1, dataframe2){ print("R script run.") A <- unserialize(as.raw(dataframe1$serialized)) # Return datasets as a Named List return(list(dataset1=dataframe1, dataset2=dataframe2)) }
Preinstalled R packages
The following preinstalled R packages are currently available:
| Package | Version |
|---|---|
| askpass | 1.1 |
| assertthat | 0.2.1 |
| backports | 1.1.4 |
| base | 3.5.1 |
| base64enc | 0.1-3 |
| BH | 1.69.0-1 |
| bindr | 0.1.1 |
| bindrcpp | 0.2.2 |
| bitops | 1.0-6 |
| boot | 1.3-22 |
| broom | 0.5.2 |
| callr | 3.2.0 |
| caret | 6.0-84 |
| caTools | 1.17.1.2 |
| cellranger | 1.1.0 |
| class | 7.3-15 |
| cli | 1.1.0 |
| clipr | 0.6.0 |
| cluster | 2.0.7-1 |
| codetools | 0.2-16 |
| colorspace | 1.4-1 |
| compiler | 3.5.1 |
| crayon | 1.3.4 |
| curl | 3.3 |
| data.table | 1.12.2 |
| datasets | 3.5.1 |
| DBI | 1.0.0 |
| dbplyr | 1.4.1 |
| digest | 0.6.19 |
| dplyr | 0.7.6 |
| e1071 | 1.7-2 |
| evaluate | 0.14 |
| fansi | 0.4.0 |
| forcats | 0.3.0 |
| foreach | 1.4.4 |
| foreign | 0.8-71 |
| fs | 1.3.1 |
| gdata | 2.18.0 |
| generics | 0.0.2 |
| ggplot2 | 3.2.0 |
| glmnet | 2.0-18 |
| glue | 1.3.1 |
| gower | 0.2.1 |
| gplots | 3.0.1.1 |
| graphics | 3.5.1 |
| grDevices | 3.5.1 |
| grid | 3.5.1 |
| gtable | 0.3.0 |
| gtools | 3.8.1 |
| haven | 2.1.0 |
| highr | 0.8 |
| hms | 0.4.2 |
| htmltools | 0.3.6 |
| httr | 1.4.0 |
| ipred | 0.9-9 |
| iterators | 1.0.10 |
| jsonlite | 1.6 |
| KernSmooth | 2.23-15 |
| knitr | 1.23 |
| labeling | 0.3 |
| lattice | 0.20-38 |
| lava | 1.6.5 |
| lazyeval | 0.2.2 |
| lubridate | 1.7.4 |
| magrittr | 1.5 |
| markdown | 1 |
| MASS | 7.3-51.4 |
| Matrix | 1.2-17 |
| methods | 3.5.1 |
| mgcv | 1.8-28 |
| mime | 0.7 |
| ModelMetrics | 1.2.2 |
| modelr | 0.1.4 |
| munsell | 0.5.0 |
| nlme | 3.1-140 |
| nnet | 7.3-12 |
| numDeriv | 2016.8-1.1 |
| openssl | 1.4 |
| parallel | 3.5.1 |
| pillar | 1.4.1 |
| pkgconfig | 2.0.2 |
| plogr | 0.2.0 |
| plyr | 1.8.4 |
| prettyunits | 1.0.2 |
| processx | 3.3.1 |
| prodlim | 2018.04.18 |
| progress | 1.2.2 |
| ps | 1.3.0 |
| purrr | 0.3.2 |
| quadprog | 1.5-7 |
| quantmod | 0.4-15 |
| R6 | 2.4.0 |
| randomForest | 4.6-14 |
| RColorBrewer | 1.1-2 |
| Rcpp | 1.0.1 |
| RcppRoll | 0.3.0 |
| readr | 1.3.1 |
| readxl | 1.3.1 |
| recipes | 0.1.5 |
| rematch | 1.0.1 |
| reprex | 0.3.0 |
| reshape2 | 1.4.3 |
| reticulate | 1.12 |
| rlang | 0.4.0 |
| rmarkdown | 1.13 |
| ROCR | 1.0-7 |
| rpart | 4.1-15 |
| rstudioapi | 0.1 |
| rvest | 0.3.4 |
| scales | 1.0.0 |
| selectr | 0.4-1 |
| spatial | 7.3-11 |
| splines | 3.5.1 |
| SQUAREM | 2017.10-1 |
| stats | 3.5.1 |
| stats4 | 3.5.1 |
| stringi | 1.4.3 |
| stringr | 1.3.1 |
| survival | 2.44-1.1 |
| sys | 3.2 |
| tcltk | 3.5.1 |
| tibble | 2.1.3 |
| tidyr | 0.8.3 |
| tidyselect | 0.2.5 |
| tidyverse | 1.2.1 |
| timeDate | 3043.102 |
| tinytex | 0.13 |
| tools | 3.5.1 |
| tseries | 0.10-47 |
| TTR | 0.23-4 |
| utf8 | 1.1.4 |
| utils | 3.5.1 |
| vctrs | 0.1.0 |
| viridisLite | 0.3.0 |
| whisker | 0.3-2 |
| withr | 2.1.2 |
| xfun | 0.8 |
| xml2 | 1.2.0 |
| xts | 0.11-2 |
| yaml | 2.2.0 |
| zeallot | 0.1.0 |
| zoo | 1.8-6 |
Next steps
See the set of components available to Azure Machine Learning.